Author: Denis Avetisyan
Researchers have developed a novel artificial intelligence model that significantly improves the reliability and clinical reasoning behind automated electrocardiogram interpretation.
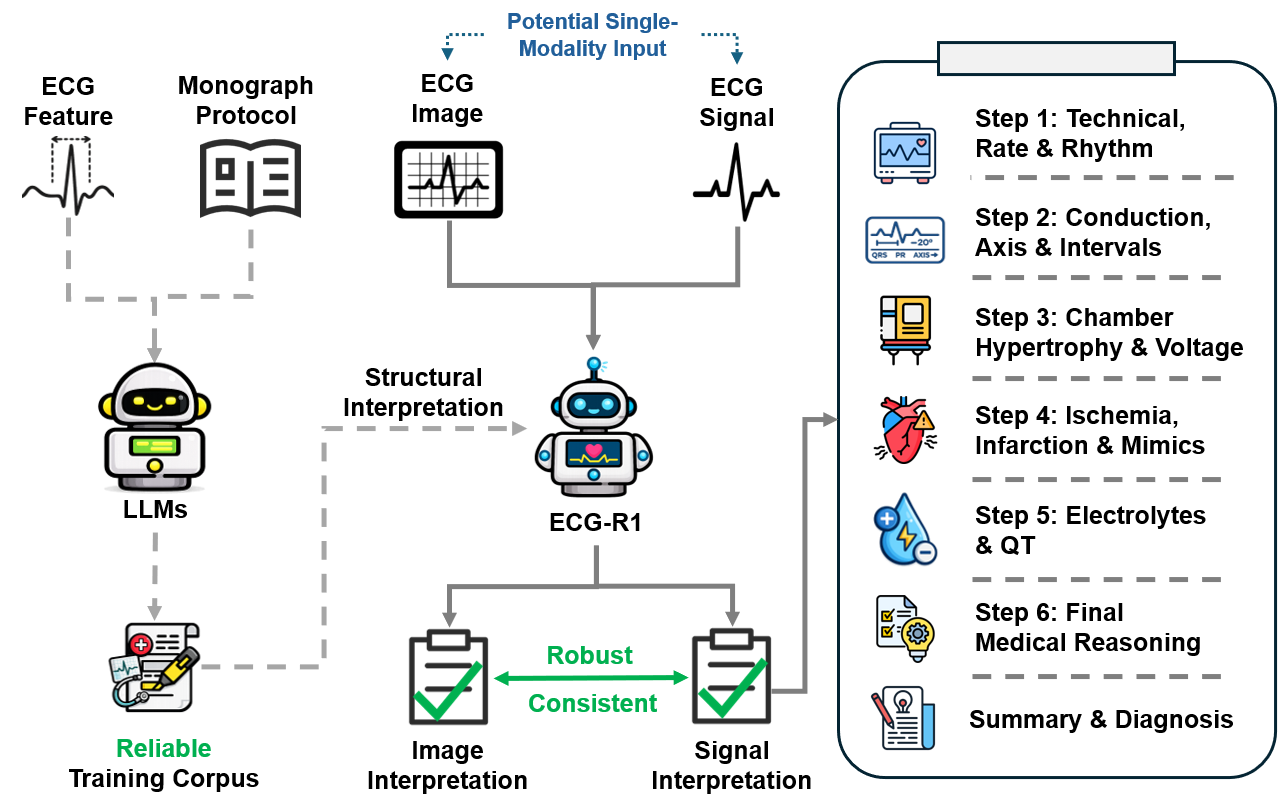
ECG-R1, a protocol-guided multimodal large language model, leverages reinforcement learning and modality dropout to mitigate hallucinations and enhance diagnostic accuracy.
Despite the critical role of electrocardiography (ECG) in clinical diagnosis, existing multimodal large language models (MLLMs) often produce plausible yet clinically incorrect interpretations. To address this unreliability, we introduce ECG-R1: Protocol-Guided and Modality-Agnostic MLLM for Reliable ECG Interpretation, a novel framework leveraging protocol-guided data generation, interleaved modality dropout, and reinforcement learning with diagnostic evidence rewards. This approach demonstrably improves both the robustness and clinical reasoning capabilities of ECG interpretation, mitigating widespread hallucinations observed in current models. Will this advancement pave the way for more trustworthy and accessible AI-driven cardiac diagnostics?
The Persistent Challenge of Accurate Cardiac Diagnosis
The accurate discernment of cardiac disease relies heavily on electrocardiogram (ECG) interpretation, a process that, despite its centrality to cardiology, presents ongoing challenges. While the ECG provides a graphical representation of the heart’s electrical activity, translating these complex waveforms into a definitive diagnosis requires significant expertise. Subtle variations, often indicative of critical conditions, can be easily overlooked or misinterpreted, leading to delayed or incorrect treatment. Factors contributing to these errors include the inherent complexity of cardiac physiology, the potential for noise and artifact in the signal, and the subjective nature of pattern recognition – even among experienced clinicians, inter-observer variability remains a concern. Consequently, enhancing the reliability of ECG interpretation is paramount to improving patient outcomes and reducing the burden of cardiovascular disease.
The interpretation of electrocardiograms (ECGs) frequently presents a challenge to even experienced clinicians due to the subtlety of many cardiac anomalies. Traditional diagnostic approaches rely heavily on pattern recognition, a process susceptible to human error when faced with complex or atypical presentations. Identifying nuanced variations – a slightly prolonged interval, a diminished voltage, or a subtle morphological change – requires a high degree of expertise and can be easily overlooked, particularly in the context of noisy data or overlapping signals. These subtle indicators often represent the earliest signs of potentially serious conditions, making their accurate detection paramount; however, the brain’s tendency to simplify information can lead to misinterpretations, highlighting the need for more objective and sensitive analytical tools to augment human expertise and improve diagnostic reliability.
The escalating prevalence of complex cardiac conditions, fueled by aging populations and lifestyle factors, is outpacing the capabilities of traditional electrocardiogram (ECG) interpretation methods. Contemporary cardiology increasingly encounters patients with comorbidities, atypical presentations, and subtle arrhythmic events that challenge diagnostic accuracy. This necessitates a shift towards more robust and reliable tools – encompassing advanced algorithms, artificial intelligence, and machine learning – capable of discerning nuanced patterns within ECG data that might be overlooked by human analysis. The demand isn’t simply for faster diagnosis, but for a heightened level of precision, minimizing false positives and negatives, and ultimately improving patient outcomes in the face of increasingly intricate cardiac pathologies.
ECG-R1: A Foundation for Rigorous Clinical Reasoning
ECG-R1 is a multimodal large language model (MLLM) developed to improve the precision and dependability of electrocardiogram (ECG) interpretation. Unlike general-purpose LLMs, ECG-R1 is specifically trained to process and understand both textual clinical data and visual ECG waveforms. This capability allows the model to integrate information from various sources – including patient history, reported symptoms, and the ECG signal itself – to provide more accurate and nuanced interpretations. The model’s architecture is designed to handle the complexities of ECG data, recognizing patterns and anomalies that might be missed by traditional analysis methods or less specialized AI systems. This targeted approach aims to reduce diagnostic errors and enhance clinical decision-making in cardiology.
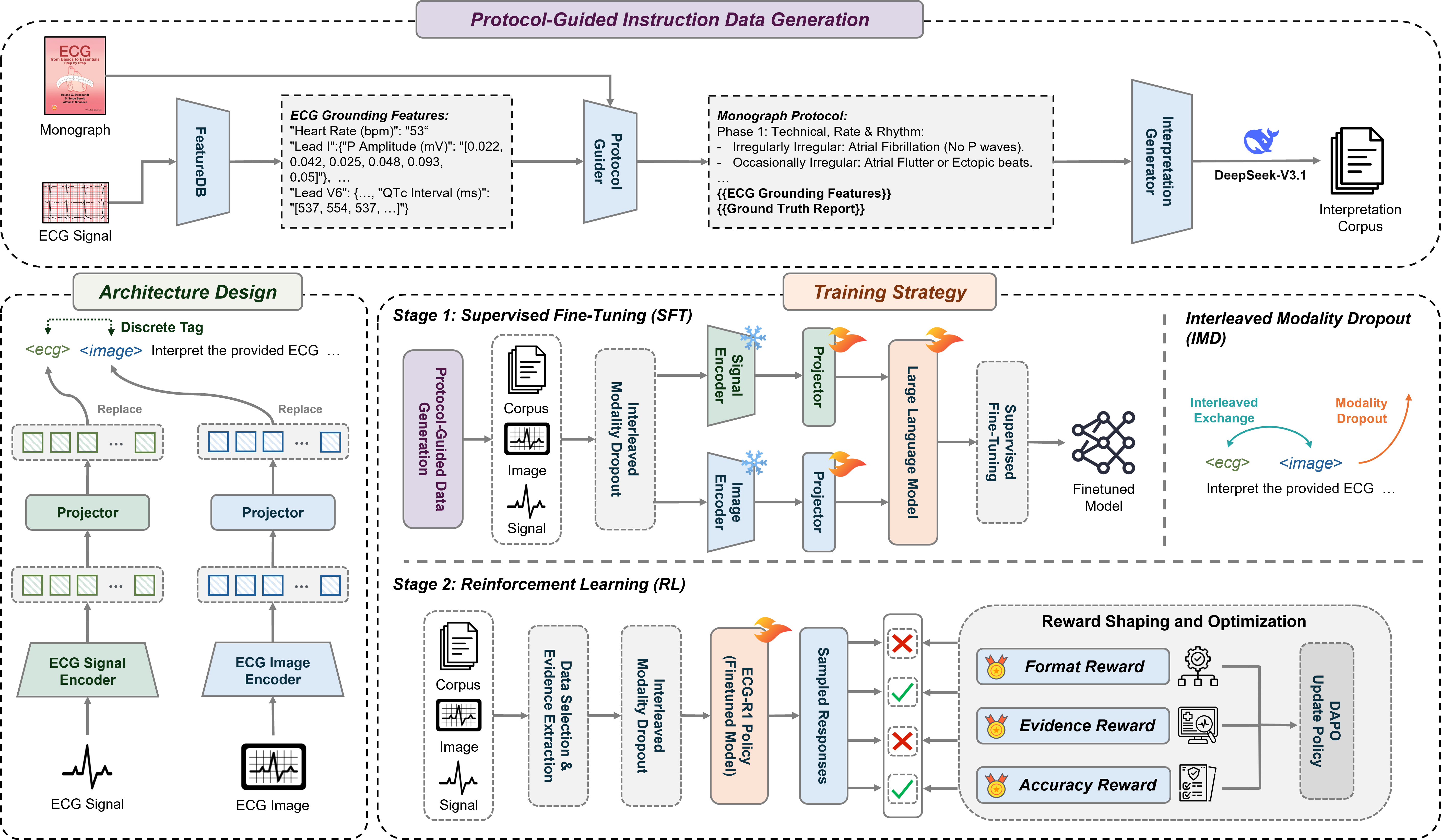
Supervised Fine-tuning (SFT) was employed to adapt a pre-trained large language model to the specific domain of electrocardiogram (ECG) data analysis. This process involves training the model on a labeled dataset of ECG readings and corresponding interpretations, allowing it to refine its existing knowledge and learn the nuances of ECG patterns. By leveraging the pre-trained model’s foundational understanding of language and data, SFT reduces the amount of data required for effective training and accelerates the learning process compared to training from scratch. The resulting model demonstrates improved performance in ECG interpretation tasks by focusing its capabilities on the complexities inherent in ECG data, such as recognizing subtle anomalies and differentiating between various cardiac conditions.
Protocol-Guided Instruction Data Generation addresses the scarcity of labeled ECG data by programmatically creating training examples based on established clinical protocols. This method utilizes information extracted from authoritative cardiology monographs – specifically, standardized interpretations of ECG characteristics and associated diagnoses – to formulate question-answer pairs. The process involves identifying key ECG features detailed within these texts and constructing prompts requesting the model to interpret those features, then automatically generating the corresponding clinical interpretation as the target answer. This automated approach ensures consistency with established medical knowledge and significantly expands the volume of high-quality, labeled data available for supervised fine-tuning of the ECG-R1 model, improving its reliability and diagnostic accuracy.
Enhancing Diagnostic Integrity Through Rigorous Training
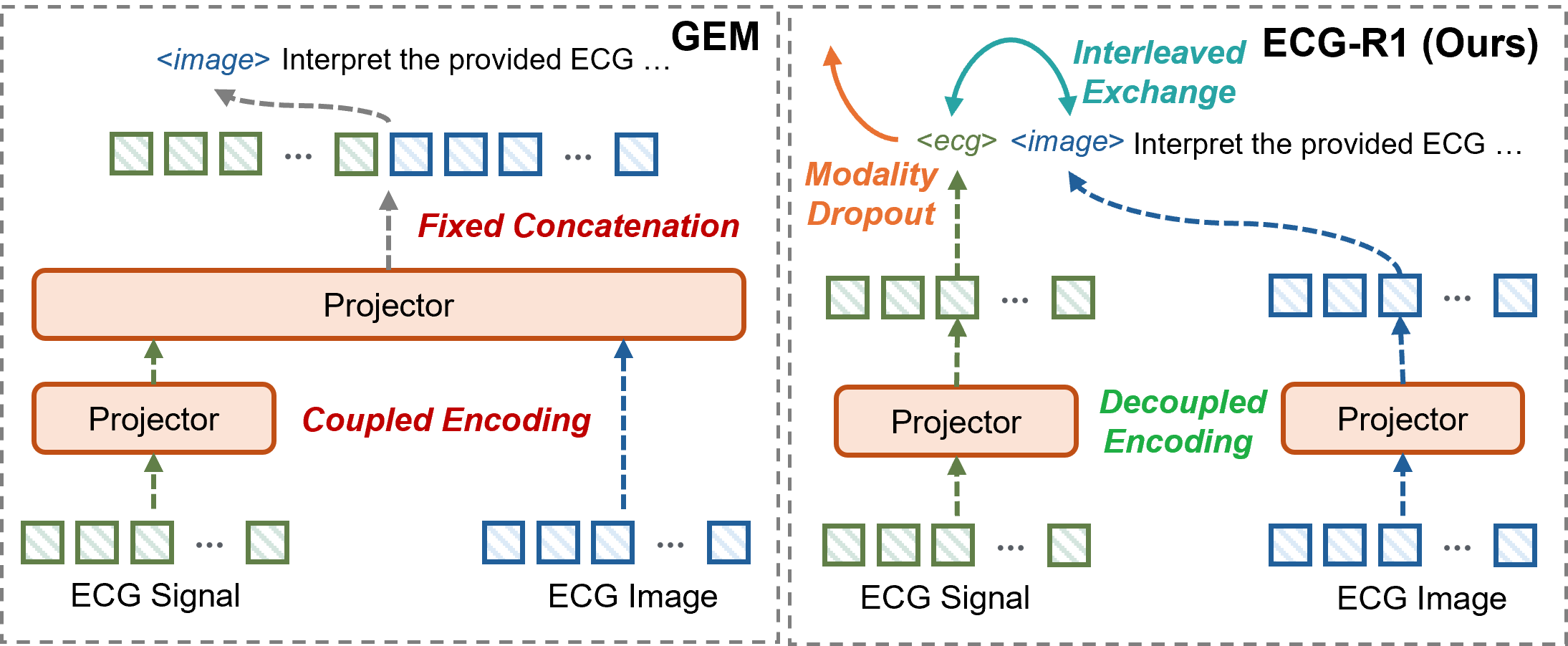
ECG-R1 utilizes Interleaved Modality Dropout during the training process to enhance model robustness. This technique randomly disables either the ECG signal input or the corresponding imaging data for individual training samples. By systematically presenting the model with incomplete data, Interleaved Modality Dropout forces it to learn representations that are less reliant on any single modality. This approach simulates real-world clinical scenarios where data acquisition may be imperfect or one data source may be unavailable, and it improves the model’s ability to generalize and maintain performance even with missing input features.
Cross-Modal Consistency, as implemented in ECG-R1, addresses potential data gaps during inference by training the model to maintain reliable interpretations even when either the ECG signal or image data is unavailable. This is achieved by intermittently masking one modality during training; the model is then forced to infer information from the remaining modality, effectively learning to correlate and compensate for missing data. The resulting model demonstrates increased resilience to incomplete inputs, as it has been explicitly trained to produce consistent outputs based on partial information, reducing the risk of inaccurate diagnoses due to data loss or sensor failure.
Reinforcement Learning (RL) is integrated into the ECG-R1 model’s training to optimize diagnostic reasoning. The RL agent receives rewards based on the alignment of its ECG interpretations with established clinical evidence; specifically, rewards are assigned when the model’s diagnostic conclusions correspond to validated clinical reasoning principles and accepted diagnostic criteria. This process refines the model’s decision-making capabilities by incentivizing interpretations that mirror expert clinical assessment. The reward function is designed to prioritize accurate diagnoses and discourage spurious correlations, effectively guiding the model toward clinically plausible reasoning pathways and improving the reliability of its outputs in complex cases.

Validation and Performance: A Paradigm Shift in Clinical Accuracy
Recent evaluations indicate that ECG-R1 represents a significant advancement in diagnostic accuracy for electrocardiogram (ECG) analysis, exceeding the performance of existing multi-modal large language models (MLLMs) like GEM. Achieving an overall diagnosis accuracy of 80.29%, ECG-R1 demonstrates a substantial improvement in reliably interpreting complex cardiac data. This heightened accuracy isn’t merely incremental; it suggests a potential for more precise and confident diagnoses, ultimately supporting clinicians in making informed decisions and improving patient outcomes. The model’s ability to consistently identify critical indicators within ECG readings positions it as a valuable tool in a variety of healthcare settings, from routine check-ups to emergency care.
Comprehensive evaluation of the ECG-R1 model was conducted utilizing the DeepSeek-V3.1-Terminus benchmark, a challenging dataset designed to assess performance in nuanced and complex diagnostic situations. Results from this rigorous testing confirm ECG-R1’s ability to accurately interpret electrocardiograms even when presented with atypical or ambiguous data, surpassing the capabilities of existing multi-modal large language models. The model consistently demonstrated a higher degree of diagnostic precision across a wide range of cardiac conditions, indicating a robust capacity for handling the intricacies inherent in real-world clinical applications and solidifying its potential as a valuable tool for healthcare professionals.
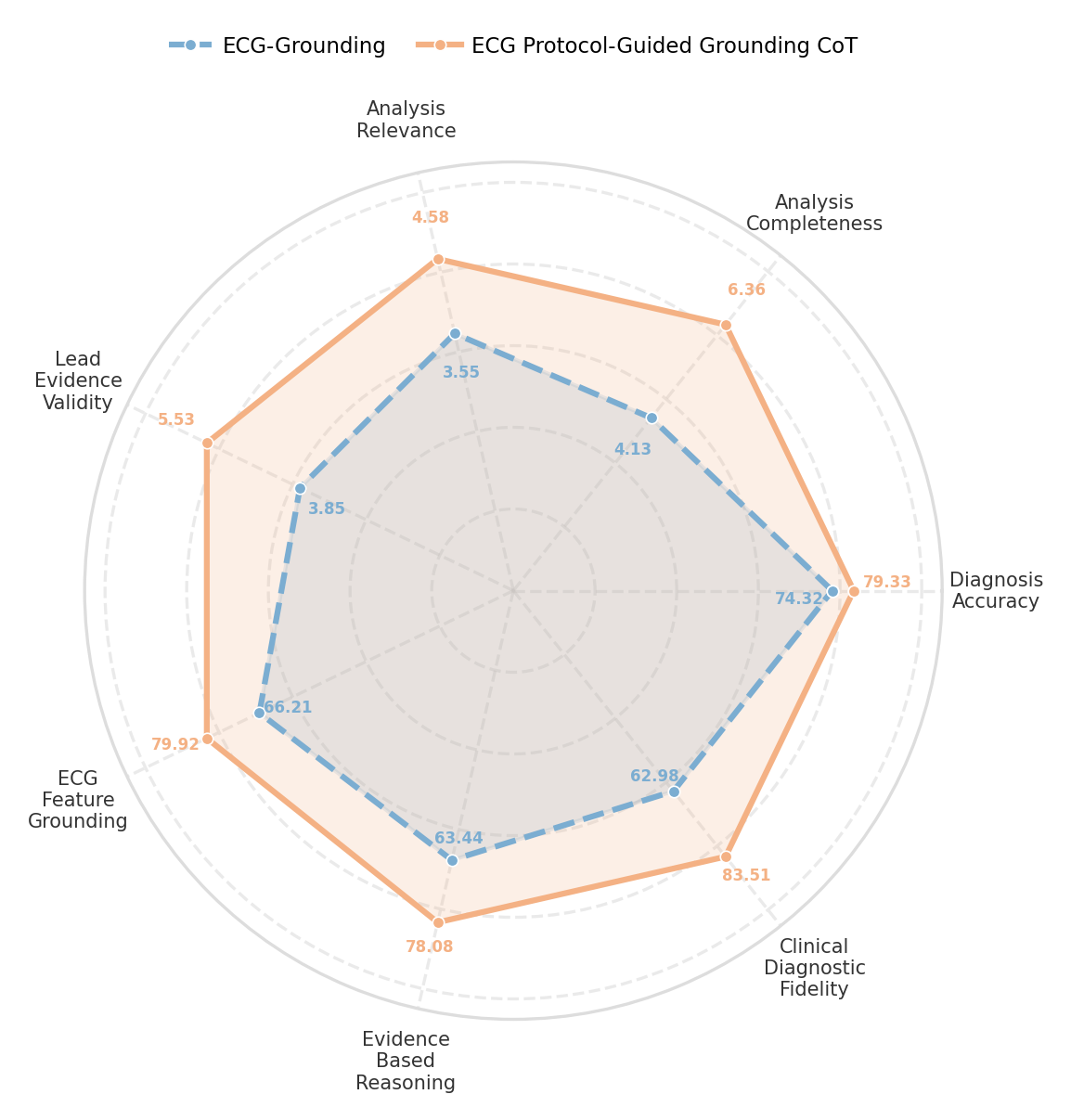
A significant advancement offered by the ECG-R1 model lies in its reduced susceptibility to “Hallucination”-a pervasive issue in medical artificial intelligence where the system generates factually incorrect or nonsensical information. Rigorous testing reveals that ECG-R1 achieves a +17.49% average absolute gain in ECG Feature Grounding when contrasted with the GEM model, indicating a substantially improved ability to anchor its diagnostic reasoning to actual, discernible features within the electrocardiogram data. This enhanced grounding not only boosts the reliability of its analyses but also minimizes the risk of producing misleading or potentially harmful interpretations, representing a crucial step toward trustworthy AI-assisted cardiac diagnosis.
ECG-R1 demonstrates a remarkable ability to integrate and reconcile information from different data sources, achieving a SBERT-Score of 0.97-a key indicator of cross-modality consistency. This signifies the model’s robust understanding and coherent interpretation of ECG data in relation to associated clinical text. Independent evaluation by cardiologists further confirms this strength, with ECG-R1 receiving an average analytical accuracy rating of 4.34 compared to 3.89 for the GEM model. This higher rating underscores ECG-R1’s superior capacity to synthesize information, leading to more reliable and clinically relevant diagnostic assessments, and suggesting a minimized risk of conflicting interpretations arising from disparate data streams.
ECG-R1 exhibits notable robustness in diagnostic scenarios where complete data isn’t available, consistently maintaining higher accuracy when relying solely on time-series ECG data compared to existing models like GEM. This resilience stems from the model’s refined architecture, allowing it to extract clinically relevant information directly from the waveform even without supplementary reports or clinical notes. Evaluations reveal a significantly reduced performance drop for ECG-R1 when operating on time-series data alone, suggesting a more effective internal representation of cardiac signals and a decreased reliance on external contextual information for accurate diagnoses. This capability is particularly valuable in real-world clinical settings where data may be incomplete or fragmented, offering a more dependable diagnostic tool across a broader range of input conditions.
The pursuit of reliable ECG interpretation, as demonstrated by ECG-R1, echoes a fundamental principle of mathematical rigor. The model’s protocol-guided data generation and reinforcement learning, focusing on diagnostic evidence, aren’t merely about achieving high accuracy-they’re about establishing a provable foundation for clinical reasoning. As Carl Friedrich Gauss stated, “I prefer a beautiful solution to a correct one.” This resonates deeply; ECG-R1 strives not simply for functional performance, but for an elegant solution built upon verifiable evidence, mitigating hallucination and bolstering robustness – a truly beautiful, and therefore trustworthy, diagnostic tool. The interleaved modality dropout is not a shortcut, but a demonstration of robustness-a method for proving the model’s ability to maintain correctness even when challenged.
Future Directions
The demonstrated efficacy of ECG-R1, while a step towards verifiable clinical reasoning in large language models, does not obviate the fundamental challenge: correlation is not causation. The model’s performance, however robust to modality dropout, remains contingent on the quality – and inherent biases – of the training data. Future work must address the provability of diagnostic inferences, shifting from empirical validation on held-out sets to formal guarantees of correctness. A crucial area for exploration is the development of symbolic reasoning layers, integrated with the LLM, capable of verifying the logical consistency of diagnostic pathways.
Furthermore, the current reward structure, predicated on diagnostic evidence, assumes a ground truth that is, in clinical practice, frequently ambiguous. The asymptotic behavior of the reinforcement learning algorithm under conditions of epistemic uncertainty warrants investigation. Can a model, trained to acknowledge its own limitations, achieve a more reliable, albeit less confident, diagnosis than one striving for absolute certainty? The exploration of Bayesian reward functions, explicitly modeling uncertainty, appears promising.
Ultimately, the true measure of progress lies not in achieving human-level performance – a moving target, at best – but in constructing models whose internal logic is transparent and verifiable. The current paradigm of opaque neural networks, while capable of impressive feats of pattern recognition, remains fundamentally unsuited to the task of medical diagnosis. A shift towards interpretable, provable AI is not merely desirable; it is ethically imperative.
Original article: https://arxiv.org/pdf/2602.04279.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- How to Unlock the Mines in Cookie Run: Kingdom
- Solo Leveling: Ranking the 6 Most Powerful Characters in the Jeju Island Arc
- Gold Rate Forecast
- Bitcoin Frenzy: The Presales That Will Make You Richer Than Your Ex’s New Partner! 💸
- Bitcoin’s Big Oopsie: Is It Time to Panic Sell? 🚨💸
- Gears of War: E-Day Returning Weapon Wish List
- Most Underrated Loot Spots On Dam Battlegrounds In ARC Raiders
- The Saddest Deaths In Demon Slayer
- How to Find & Evolve Cleffa in Pokemon Legends Z-A
- Rocket League: Best Controller Bindings
2026-02-06 06:55