Author: Denis Avetisyan
Researchers have developed a novel contrastive learning framework that enhances ECG analysis by focusing on both the rhythm and individual characteristics of each heartbeat.
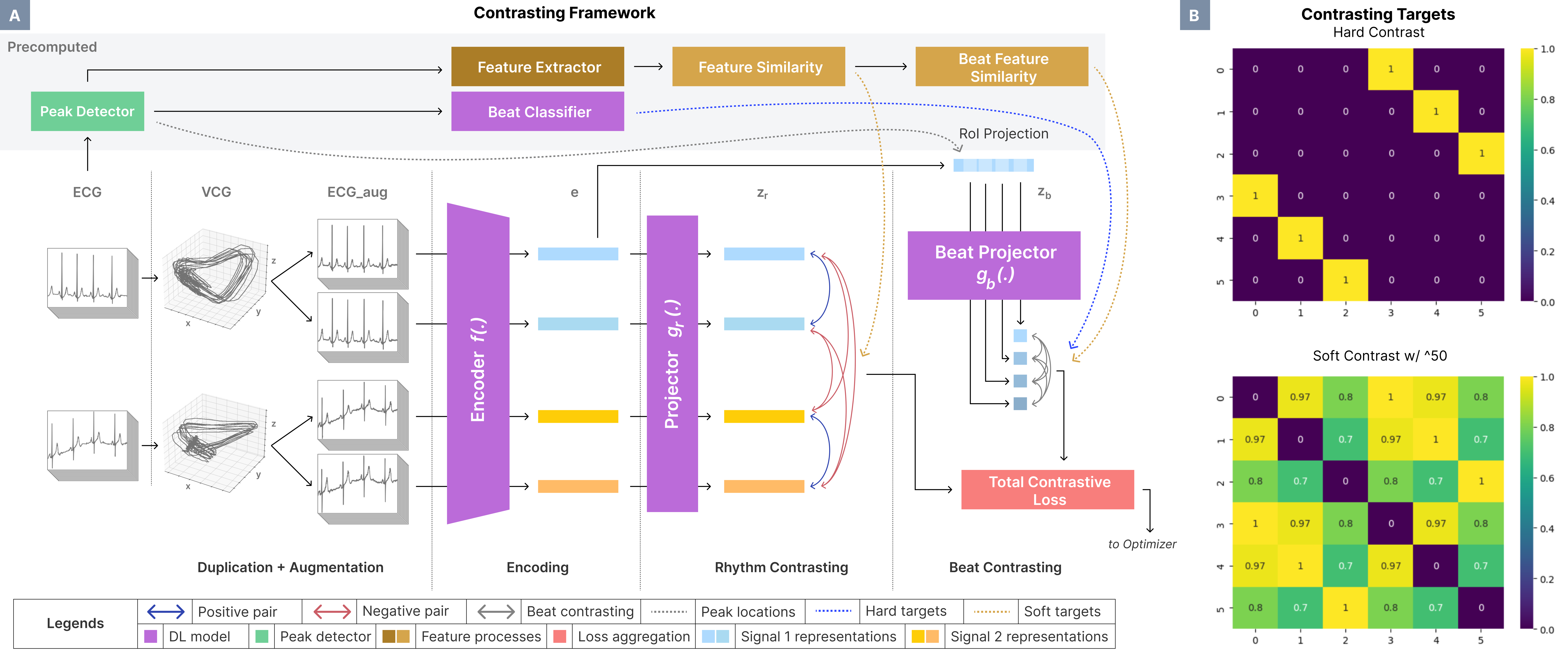
This work introduces Beat-SSL, a semi-supervised learning method leveraging heartbeat-level contrastive learning with soft targets for improved multilabel classification and ECG wave segmentation.
Obtaining sufficient labeled data remains a significant challenge in developing robust electrocardiogram (ECG) analysis models. This limitation motivates the work presented in ‘Beat-ssl: Capturing Local ECG Morphology through Heartbeat-level Contrastive Learning with Soft Targets’, which introduces a novel contrastive learning framework leveraging both rhythm-level and heartbeat-level information with soft target assignments. The proposed Beat-SSL achieves state-of-the-art performance in both multilabel classification and ECG segmentation, surpassing existing methods including a prominent ECG foundation model. By effectively capturing local ECG morphology, can this approach pave the way for more accurate and efficient automated cardiac diagnostics?
The ECG Data Deluge: Why More Data Isn’t Always Better
Effective electrocardiogram (ECG) analysis using traditional machine learning techniques is fundamentally limited by the need for vast quantities of meticulously labeled data – a considerable obstacle within real-world clinical settings. The process of accurately annotating ECG signals, identifying subtle arrhythmias or ischemic events, demands highly trained cardiologists and is exceptionally time-consuming and expensive. This reliance on extensive labeling creates a significant bottleneck, hindering the widespread adoption of automated ECG diagnostic tools and impeding research efforts. Consequently, the development of algorithms often stalls due to the scarcity of readily available, high-quality datasets, particularly for less common cardiac conditions, effectively restricting the potential for personalized and proactive cardiovascular care.
Despite the initial enthusiasm surrounding deep learning’s potential in electrocardiogram (ECG) analysis, these models frequently encounter difficulties when applied to unseen data. ECG signals exhibit substantial variability, not only between individuals-influenced by age, sex, and pre-existing conditions-but also within the same patient over time, due to factors like body position, breathing, and even minor muscle movements. This inherent complexity means a model trained on one dataset may perform poorly when exposed to ECGs acquired with different equipment, sampling rates, or patient populations. The subtle nuances within these waveforms, crucial for accurate diagnosis, are often lost in the generalization process, requiring innovative techniques to improve the robustness and adaptability of deep learning algorithms for reliable clinical application.
Electrocardiogram (ECG) signals present a distinct analytical challenge, differing fundamentally from typical time series or image data. Unlike the relatively stationary characteristics of many datasets used in those fields, ECG signals are inherently non-stationary and exhibit complex morphology influenced by physiological factors and noise. Standard time series analysis often struggles with the signal’s varying frequency content and transient events, while computer vision techniques, designed for spatially correlated data, fail to adequately capture the temporal dependencies crucial to ECG interpretation. Consequently, researchers are increasingly focused on developing specialized algorithms – incorporating domain knowledge of cardiac electrophysiology – and leveraging novel deep learning architectures tailored to the unique properties of ECG data, such as recurrent neural networks capable of modeling sequential information and convolutional networks adapted for one-dimensional signal processing. These approaches aim to move beyond generalized methods and unlock the full potential of ECG analysis for improved diagnostics and patient care.
Unlabeled Data to the Rescue: A Self-Supervised Approach
Contrastive learning (CL) provides a self-supervised approach to model pretraining by enabling models to learn representations from unlabeled electrocardiogram (ECG) data. This is achieved by training the model to maximize the similarity between representations of augmented versions of the same ECG sample (positive pairs) while minimizing the similarity between representations of different ECG samples (negative pairs). The process does not require manual annotations, allowing for the utilization of large volumes of readily available unlabeled ECG data. By learning to discern subtle differences and similarities in ECG waveforms, the model develops a robust feature space that can then be leveraged for downstream tasks with limited labeled data, ultimately improving performance and reducing the reliance on costly and time-consuming manual annotation efforts.
Contrastive learning (CL) pretraining enables the development of foundation models that significantly reduce the reliance on large, labeled ECG datasets for downstream tasks. By learning robust feature representations from unlabeled data, these models require substantially less labeled data during the fine-tuning stage to achieve high diagnostic accuracy and improved generalization performance. This is particularly valuable in cardiology, where obtaining extensive, expertly annotated ECG data is often costly and time-consuming. The reduced data requirement also mitigates the risk of overfitting to limited labeled examples, leading to more reliable and clinically applicable models.
Heartbeat-level contrasting in ECG analysis involves treating each individual heartbeat as a distinct data point for comparison. This methodology moves beyond analyzing entire ECG recordings and instead concentrates on the morphological characteristics of each beat – specifically, the P-wave, QRS complex, and T-wave. By contrasting these beat morphologies, the model learns to identify subtle variations indicative of cardiac abnormalities, such as arrhythmias or ischemia, that might be missed when considering only broader ECG patterns. This fine-grained analysis is crucial because even minor alterations in beat morphology can be clinically significant, and accurately detecting these nuances requires a high degree of discriminatory power within the pretraining phase of the model.
Defining Similarity: Moving Beyond Binary Labels
Traditional machine learning approaches often rely on assigning discrete, or ‘hard’, labels to electrocardiogram (ECG) beats – categorizing them as belonging to specific arrhythmia classes. In contrast, this methodology generates ‘soft targets’ by quantifying the degree of similarity between ECG beat features. Rather than a binary classification, this produces a continuous value representing relatedness, allowing for nuanced distinctions between beat morphologies. This is achieved by calculating a similarity score based on the feature vectors of individual beats, effectively capturing the degree to which they resemble each other, and providing a richer representation of beat relationships than simple categorical labels.
Exponentiation of similarity scores is implemented to amplify the distinction between highly similar ECG beats, enabling the model to prioritize subtle morphological variations. This process increases the magnitude of already high similarity values, effectively sharpening the contrast used during training. By focusing on these nuanced differences, the model is encouraged to learn features that are clinically relevant for arrhythmia detection and classification, rather than being dominated by more prominent, but potentially less informative, beat characteristics. The resulting scaled values are then used as targets in the contrastive loss function, guiding the model’s feature embedding process.
Soft targets are generated through pseudo-labeling, a process utilizing a pre-trained beat-classifier. This classifier was trained on the MIT-BIH Arrhythmia Database, a widely-used and clinically validated resource for arrhythmia detection. The output of this classifier, representing predicted beat categories, serves as the basis for assigning similarity scores between ECG beats. This approach ensures that the generated soft targets reflect clinically relevant distinctions between normal and abnormal heartbeats, as defined by expert annotations within the MIT-BIH dataset. The use of a pre-trained, clinically-validated classifier is critical for grounding the similarity metric in established medical knowledge.
The Normalized Temperature-scaled Cross Entropy loss (NT-Xent) functions by maximizing the agreement between embeddings of similar ECG beat morphologies and minimizing agreement between dissimilar morphologies. This is achieved by constructing positive and negative pairs; positive pairs consist of embeddings derived from beats identified as similar via the soft target methodology, while negative pairs are formed from dissimilar beats. The loss function calculates a contrastive score based on cosine similarity between these embedding pairs, and utilizes a temperature parameter to scale the similarity scores, sharpening the distinction between positive and negative examples. The resulting gradient updates during training effectively push similar beat embeddings closer together in the feature space and push dissimilar embeddings further apart, enabling the model to learn robust representations of beat morphology.
Beyond Beats: Capturing the Full Context of Cardiac Signals
Electrocardiogram (ECG) signals, while seemingly straightforward recordings of heart activity, contain intricate patterns that demand sophisticated analysis. Recent advancements leverage self-supervised learning techniques, notably Wave2vec and Contrastive Multi-segment Coding (CMSC), to effectively capture both the immediate, localized features – such as the precise shape of a QRS complex – and the broader, global context of the entire cardiac cycle. Wave2vec, originally developed for speech recognition, learns robust representations by predicting masked portions of the ECG signal, forcing the model to understand the relationships between adjacent waveform segments. Complementing this, CMSC enhances contextual awareness by contrasting different segments of the ECG, encouraging the model to discern subtle but clinically significant variations. This dual approach enables a more nuanced interpretation of ECG data, moving beyond simple beat detection to a comprehensive understanding of cardiac function and pathology.
The 3KG model enhances data diversity and system resilience through strategic vectorcardiography (VCG) transformations. By converting standard electrocardiogram (ECG) signals into their corresponding VCG representations, the model effectively expands the training dataset without requiring additional patient data. This process generates synthetic variations of existing ECGs, exposing the system to a wider range of physiological conditions and improving its ability to generalize to unseen data. The VCG transformation acts as a form of data augmentation, increasing the robustness of the model against noise and variations in signal quality, ultimately leading to improved performance in downstream tasks like multi-label arrhythmia classification and precise ECG wave segmentation.
The ability to interpret electrocardiogram (ECG) signals relies heavily on understanding the context within the data, and recent advancements demonstrate the critical impact of this contextual awareness on diagnostic accuracy. Specifically, employing techniques that capture both local and global patterns within the ECG has yielded significant improvements in downstream tasks; for instance, a multi-label classification challenge, termed the Superdiagnostic Task, achieved a robust F1-score of 93%. This contextual understanding also proves invaluable for precise ECG segmentation, as evidenced by results obtained from the LUDB Dataset, showcasing the potential to unlock more detailed and accurate interpretations of cardiac activity and ultimately, enhance patient care.
Recent advancements in electrocardiogram (ECG) analysis demonstrate a significant leap in the precision of waveform segmentation. A novel approach to contextual understanding within ECG signals has yielded a 4% improvement in the Dice score – a key metric for evaluating segmentation accuracy – when compared to existing state-of-the-art methods. This enhancement indicates a superior ability to discern the subtle, yet critical, features defining each wave, achieved through effective capture of local contextual information. The improvement isn’t merely statistical; it represents a more refined understanding of the signal’s morphology, potentially leading to earlier and more accurate diagnoses of cardiac abnormalities through precise wave delineation.
A significant advantage of this approach lies in its data efficiency; the method achieves comparable, and in some cases superior, performance with substantially less training data. Specifically, the system requires approximately 700,000 data points for pretraining, representing a 31.8-fold reduction compared to the data demands of the ECG-FM method. This minimized data requirement not only lowers computational costs but also broadens accessibility, allowing for effective model training even with limited datasets-a critical factor in medical applications where data acquisition can be challenging and expensive.
The pursuit of elegant models, as demonstrated in this work on heartbeat-level contrastive learning, inevitably courts eventual compromise. This paper’s meticulous approach to ECG analysis – leveraging both rhythm and heartbeat levels with soft and hard contrasting techniques – feels less like a solution and more like a beautifully engineered delay of the inevitable. As John McCarthy observed, “It is perhaps a bit optimistic to think that machines will ever be able to understand human language and thought.” The same applies here; the model may achieve state-of-the-art performance in multilabel classification and ECG wave segmentation now, but production data will always find the edge cases, the subtle anomalies, that reveal the abstraction’s limits. It dies beautifully, this model, but it will die.
What’s Next?
This work, predictably, introduces a new complexity to a field already drowning in edge cases. The pursuit of ‘foundation models’ for ECG analysis, while logically appealing, merely shifts the burden of feature engineering. Any architecture capable of capturing ‘local ECG morphology’ will inevitably require more data – and the endless, Sisyphean task of labeling it. The current reliance on both ‘hard’ and ‘soft’ contrasting techniques feels less like an elegant solution and more like an admission that the signal is fundamentally ambiguous.
The performance gains achieved through heartbeat-level contrastive learning are unlikely to remain unassailable. Production environments will reveal unseen pathologies, artifacts, and the inherent variability of human physiology. The reported multilabel classification success will be challenged by datasets that reflect real-world clinical heterogeneity-a chaos this framework, for all its cleverness, has yet to encounter.
Future iterations will undoubtedly focus on reducing the labeling requirements-a noble goal. However, it’s reasonable to suspect that each simplification will introduce another layer of abstraction, further distancing the model from the raw, messy reality of the cardiac cycle. Documentation, of course, will remain a myth invented by managers. CI is the new temple – and the prayers for unbroken pipelines will only grow louder.
Original article: https://arxiv.org/pdf/2601.16147.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- Enshrouded: Giant Critter Scales Location
- Best Finishers In WWE 2K25
- Top 8 UFC 5 Perks Every Fighter Should Use
- How to Unlock & Visit Town Square in Cookie Run: Kingdom
- Best Anime Cyborgs
- All Carcadia Burn ECHO Log Locations in Borderlands 4
- All Shrine Climb Locations in Ghost of Yotei
- Poppy Playtime 5: Battery Locations & Locker Code for Huggy Escape Room
- Best ARs in BF6
- Top 10 Must-Watch Isekai Anime on Crunchyroll Revealed!
2026-01-25 12:12