Author: Denis Avetisyan
A new approach to knowledge representation allows question answering systems to dynamically adapt to evolving information and express the certainty of their responses.
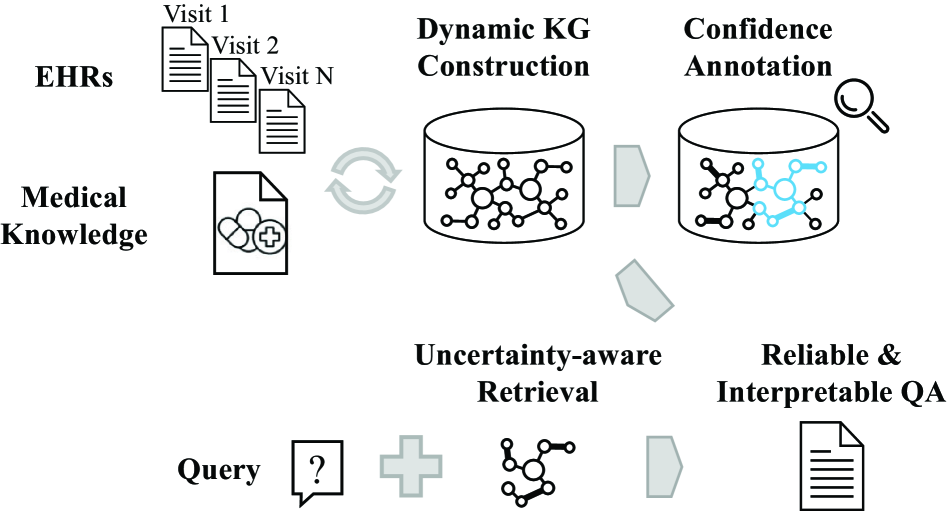
This review details uncertainty-aware dynamic knowledge graphs and their application to improving the reliability of question answering, particularly in complex domains like clinical reasoning.
Despite the increasing deployment of question answering (QA) systems, their reliability remains challenged by incomplete, noisy, or uncertain evidence-a critical limitation particularly in dynamic fields like healthcare. This paper introduces ‘Uncertainty-Aware Dynamic Knowledge Graphs for Reliable Question Answering’, a framework that addresses this challenge by constructing evolving knowledge graphs with associated confidence scores. Our system demonstrably enhances QA robustness and interpretability through dynamic graph exploration and confidence-annotated reasoning. Could this approach unlock more trustworthy AI-driven insights in high-stakes applications demanding both accuracy and transparency?
The Weight of Information: A System Under Strain
Healthcare generates vast quantities of data – from electronic health records and genomic sequencing to wearable sensor readings and imaging scans – yet transforming this wealth of information into practical, actionable insights presents a significant challenge. The sheer volume often overwhelms clinicians, obscuring critical patterns and hindering effective decision-making. Data is frequently siloed across disparate systems, lacking interoperability and standardization, which further complicates analysis. Moreover, much of the data is unstructured – existing as free-text notes or complex images – requiring sophisticated natural language processing and computer vision techniques to extract meaningful content. Consequently, despite unprecedented access to patient information, healthcare professionals frequently struggle to synthesize it efficiently, potentially leading to delayed diagnoses, suboptimal treatments, and increased costs.
The modern healthcare landscape presents clinicians with an overwhelming influx of patient data, frequently exceeding their cognitive capacity for effective processing. This cognitive overload isn’t simply a matter of volume; existing electronic health record systems often deliver information in a disorganized or untimely manner, hindering rather than assisting in critical decision-making. Studies reveal that physicians spend a significant portion of their time navigating these systems, searching for pertinent details instead of focusing on patient assessment and care. Consequently, relevant clinical insights can be obscured within the sheer mass of data, increasing the risk of diagnostic errors and suboptimal treatment plans. The challenge lies not just in collecting more data, but in developing systems that intelligently filter, synthesize, and present information in a format that supports, rather than strains, the clinician’s cognitive resources.
The escalating intricacy of modern medical cases necessitates a shift beyond simplistic diagnostic models. Traditional knowledge representation, often relying on rigid taxonomies and rule-based systems, struggles to capture the subtle interplay of factors defining individual patient presentations. Current approaches frequently fail to account for the probabilistic nature of disease, the influence of co-morbidities, and the variability in patient responses to treatment. Consequently, research is increasingly focused on developing more nuanced methods-such as Bayesian networks, machine learning algorithms, and knowledge graphs-capable of representing uncertainty, capturing contextual information, and facilitating reasoning under conditions of incomplete or ambiguous data. These advanced systems aim to move beyond merely identifying potential diagnoses to providing clinicians with a richer, more holistic understanding of each patient’s unique clinical profile, ultimately supporting more informed and effective decision-making.
Building the Foundation: A Living Map of Knowledge
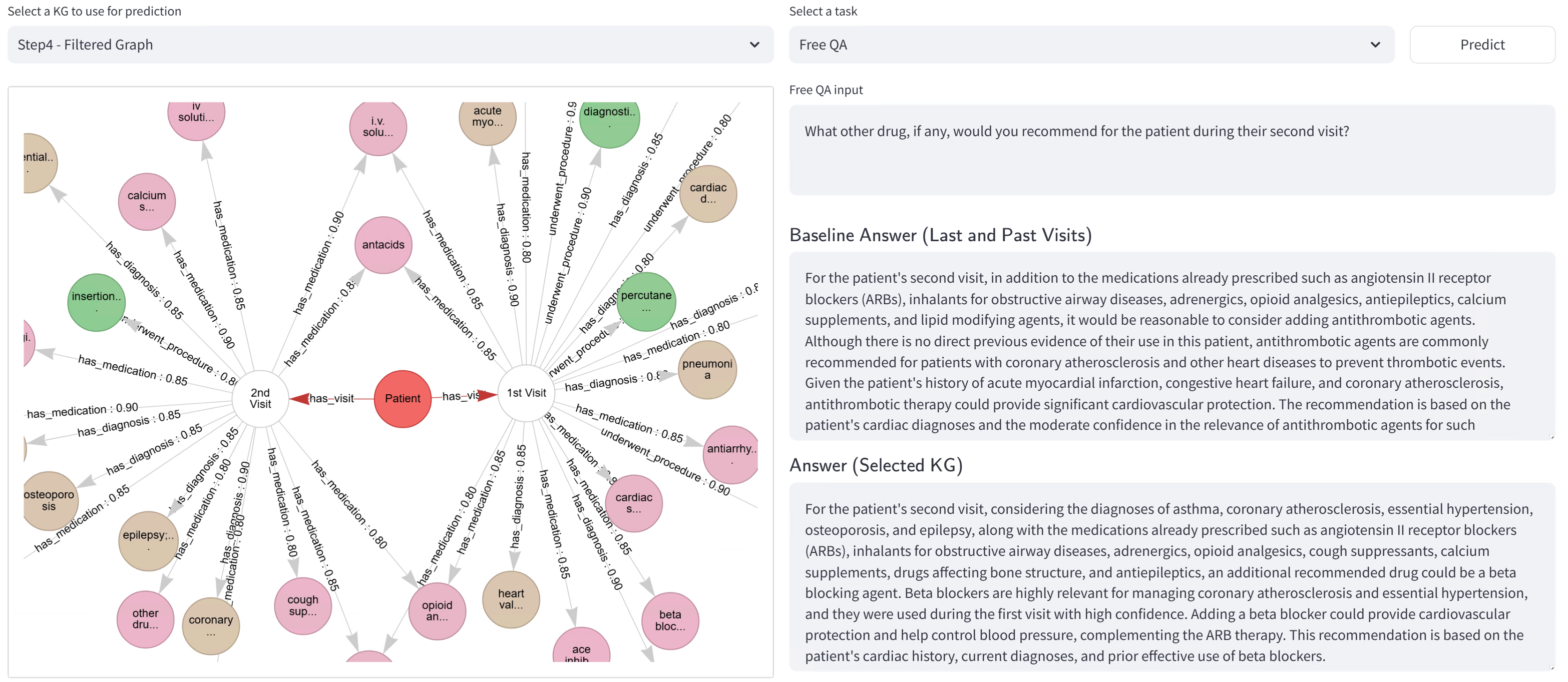
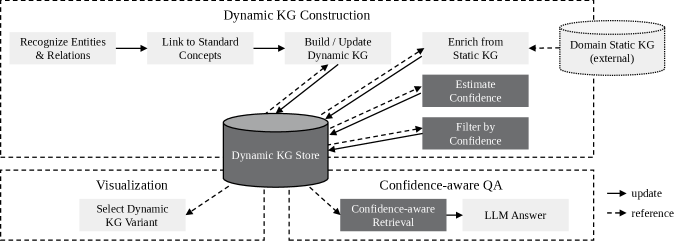
The proposed system leverages a Dynamic Knowledge Graph to represent and integrate patient data in a continuously updating format. Unlike static knowledge bases, this graph is designed to accommodate new information from various sources – including electronic health records, genomic data, and research publications – without requiring substantial restructuring. This adaptability is achieved through the graph’s inherent flexibility in adding new nodes (concepts) and edges (relationships) as data becomes available. The dynamic nature of the graph facilitates real-time updates and allows the system to reflect the most current understanding of a patient’s condition, supporting more informed clinical decision-making and personalized treatment plans.
The Dynamic Knowledge Graph represents clinical information as a network of interconnected concepts using subject-predicate-object triples. Each triple defines a relationship; for example, “Patient X has symptom Fever” or “Drug Y treats Disease Z”. The subject and object represent entities – patients, diseases, drugs, genes, etc. – while the predicate defines the specific relationship between them. This structure allows for complex relationships to be modeled and facilitates traversal of the graph to infer new knowledge and connections between clinical data points, creating a richly interconnected and informative network.
Neo4j, a native graph database, was selected as the foundational data layer due to its capacity to efficiently manage and query highly interconnected data. Unlike relational databases which require complex joins to navigate relationships, Neo4j stores relationships as first-class citizens, enabling traversals and pattern matching with significantly reduced latency. This architecture is crucial for a Dynamic Knowledge Graph where clinical concepts are extensively linked. Furthermore, Neo4j’s scalability, achieved through clustering and replication, allows the system to accommodate increasing volumes of patient data and maintain consistent performance. Its Cypher query language provides a declarative method for expressing complex relationships, simplifying data retrieval and analysis compared to SQL-based approaches.
Confidence scoring within the Dynamic Knowledge Graph assigns numerical values to relationships and concepts based on the provenance and supporting evidence of the data. This scoring system utilizes factors such as data source reliability, frequency of occurrence in validated datasets, and the consensus among multiple clinical guidelines to determine a confidence level. Scores are normalized to a consistent scale, allowing for quantitative comparison of information reliability. Lower confidence scores flag potentially uncertain data, triggering validation workflows or indicating a need for further investigation, while higher scores denote well-established and trusted clinical knowledge. The implementation employs a Bayesian approach to update confidence scores as new data is integrated, dynamically reflecting the evolving certainty of the information within the graph.
Personalizing the View: From Data to Patient Story
The system moves beyond a generalized knowledge base by constructing a Personalized Knowledge Graph (PKG) derived from the core Dynamic Knowledge Graph. This PKG integrates individual patient data – including medical history, diagnoses, medications, allergies, and lab results – to create a unique representation of each patient’s clinical profile. This individualized data is linked to relevant concepts within the Dynamic Knowledge Graph, enabling the system to contextualize information specifically to the patient. The resulting PKG facilitates retrieval of highly relevant data points when responding to clinical queries, thereby improving the accuracy and applicability of generated insights.
Retrieval-Augmented Generation (RAG) is a technique that integrates information retrieval with the capabilities of generative language models. Traditional generative models can sometimes produce inaccurate or hallucinated content, particularly when dealing with specialized or evolving knowledge domains. RAG mitigates this by first retrieving relevant documents or data snippets from a knowledge source-such as a vector database containing patient records or medical literature-based on a user’s query. These retrieved passages are then provided as context to the generative model, guiding its response and grounding it in factual information. This approach allows the model to leverage both the broad knowledge encoded in its parameters and the specificity of the retrieved data, resulting in more accurate, reliable, and contextually relevant outputs.
The system addresses complex clinical queries by integrating information retrieval with generative AI. Initially, relevant data is retrieved from the Personalized Knowledge Graph based on the specifics of the question and the patient’s history. This retrieved information is then provided as context to the GPT-4.1-mini Large Language Model, which generates a response grounded in both established medical knowledge and individualized patient data. This process ensures responses are not only comprehensive and coherent but also directly applicable to the clinical context, increasing the reliability and relevance of the information provided to healthcare professionals.
The system leverages the GPT-4.1-mini Large Language Model (LLM) to synthesize retrieved knowledge into comprehensive and understandable responses. GPT-4.1-mini is employed for its balance of computational efficiency and text generation quality, enabling the system to produce coherent clinical explanations and answers. Following the retrieval of relevant data from the Personalized Knowledge Graph, the LLM processes this information and generates text that is contextually appropriate and informative for the specific clinical query. The model’s architecture allows it to maintain semantic consistency and produce responses that are both accurate and readily interpretable by healthcare professionals.
Validation and Implementation: A System Takes Shape
The Question Answering System’s architecture utilizes FastAPI, a modern, fast (high-performance), web framework, for the backend application programming interface (API). FastAPI was selected for its efficiency in handling asynchronous requests and automatic data validation. The system’s user interface is constructed with Streamlit, an open-source Python library that simplifies the creation and deployment of interactive web applications. Streamlit enables rapid prototyping and facilitates the display of question answering results in a user-friendly format without requiring front-end web development expertise. This combination allows for a scalable and easily deployable system capable of handling user queries and presenting responses through a web browser.
System performance was evaluated using the MIMIC-III (Medical Information Mart for Intensive Care III) dataset, a publicly available critical care database comprising data associated with over 40,000 patients admitted to intensive care units. The selected task was mortality prediction, wherein the system was trained to predict in-hospital mortality based on patient features recorded during their stay. The MIMIC-III dataset provided a standardized and comprehensive source of clinical data, allowing for rigorous evaluation of the Question Answering System’s predictive capabilities in a realistic healthcare setting. Data utilized included demographics, vital signs, laboratory results, medications, and diagnoses.
The performance of the Question Answering System was quantitatively assessed using two key metrics: Area Under the Receiver Operating Characteristic Curve (AUROC) and Area Under the Precision-Recall Curve (AUPRC). AUROC, representing the probability that the model ranks a randomly chosen positive instance higher than a randomly chosen negative instance, provides a general measure of discriminatory power. AUPRC, calculated by summing the rectangular areas under the precision-recall curve, is particularly useful when dealing with imbalanced datasets, as is common in medical applications like mortality prediction. High values for both AUROC and AUPRC indicate robust predictive capabilities and the system’s ability to reliably differentiate between positive and negative cases.
Performance evaluation of the Question Answering System on the Mortality Prediction Task, using the MIMIC-III dataset, yielded statistically significant improvements over a baseline approach. Specifically, the system achieved up to an 27.4% improvement in Area Under the Precision-Recall Curve (AUPRC) and an 18.1% improvement in Area Under the Receiver Operating Characteristic Curve (AUROC). These metrics, AUPRC and AUROC , quantify the system’s ability to correctly identify positive cases while minimizing false positive rates, indicating robust predictive capability and a substantial gain in performance compared to the established baseline.
The Question Answering System incorporates multiple safeguards to ensure compliance with the Health Insurance Portability and Accountability Act (HIPAA). These include data encryption both in transit and at rest, utilizing Transport Layer Security (TLS) for secure communication and AES-256 encryption for stored data. Access controls are implemented based on the principle of least privilege, restricting data access to authorized personnel only. Audit trails are maintained to log all data access and modification events, facilitating accountability and investigation. Furthermore, the system is designed to support data de-identification and anonymization techniques, enabling the use of patient data for research and development purposes while protecting individual privacy.
Beyond Prediction: A System with Expanding Horizons
The developed system addresses a critical need in modern healthcare: the effective integration and analysis of increasingly complex clinical datasets. By employing a modular architecture and standardized data formats, the framework isn’t limited by the volume or variety of information – encompassing patient history, genomic data, imaging results, and real-time monitoring. This scalability is paramount, allowing the system to adapt as data sources expand and evolve. More importantly, this integration isn’t merely about storage; it actively facilitates knowledge discovery through advanced analytics and machine learning algorithms, enabling researchers and clinicians to identify patterns, predict outcomes, and ultimately personalize patient care with greater precision and efficiency. The system’s design prioritizes not just the ‘what’ of the data, but the ‘how’ of transforming it into actionable insights.
Advancing this predictive framework necessitates the integration of robust uncertainty modeling techniques. Current machine learning models often provide point predictions without adequately conveying the confidence level associated with those predictions, which can be problematic in high-stakes clinical settings. Future development will focus on quantifying prediction uncertainty – not just what a model predicts, but how sure it is – using methods like Bayesian neural networks or ensemble techniques. Communicating this uncertainty effectively to clinicians is paramount; visualizations displaying prediction intervals or probabilistic forecasts can enable more informed decision-making and foster appropriate trust in the system’s outputs. By explicitly representing the limits of its knowledge, the framework moves beyond simple prediction and towards a more transparent and reliable clinical decision support tool.
The adaptable nature of this framework positions it for substantial expansion beyond its initial scope, promising benefits across numerous clinical domains. While presently focused on predictive capabilities, the system’s architecture readily lends itself to supporting more complex tasks like differential diagnosis, where nuanced data integration is crucial for identifying the most likely condition from a range of possibilities. Furthermore, the framework’s ability to synthesize patient-specific information could be directly applied to personalized treatment planning, allowing clinicians to evaluate various therapeutic options and predict individual responses with greater accuracy. This extensibility isn’t limited to specific disease areas; the core principles of data harmonization and predictive modeling are broadly applicable, suggesting potential uses in preventative care, resource allocation, and even public health initiatives, ultimately fostering a more proactive and data-driven approach to healthcare.
The potential for improved clinical understanding and collaborative decision-making is significantly amplified through interactive visualization tools like pyvis. These platforms move beyond static reports, allowing clinicians to dynamically explore complex datasets and observe relationships between variables in real-time. By visually representing patient data – encompassing everything from genomic information to treatment response – pyvis facilitates a more intuitive grasp of individual cases and broader population trends. This enhanced clarity not only supports more accurate diagnoses and personalized treatment plans, but also fosters a shared mental model amongst healthcare teams, streamlining communication and promoting more effective collaboration. Ultimately, such tools promise to bridge the gap between data analysis and clinical practice, empowering healthcare professionals to leverage the full potential of complex information for improved patient outcomes.
The pursuit of reliable question answering, as detailed in this work, isn’t about imposing order, but acknowledging inherent instability. This system, constructing dynamic knowledge graphs with confidence scoring, doesn’t prevent decay-it measures it. It anticipates the inevitable erosion of information, much like a seasoned cartographer charts shifting coastlines. As Henri Poincaré observed, “Mathematics is the art of giving reasons, even to those who do not understand.” This echoes within the graph’s ability to provide not just answers, but a degree of belief in those answers – a reasoned response even when facing incomplete or evolving data, recognizing that perfect architectural solutions are merely temporary illusions in the face of entropy.
What Lies Ahead?
This work, like all attempts to formalize understanding, builds a map, forgetting the territory is forever shifting. The incorporation of confidence scoring into dynamic knowledge graphs is not a solution to uncertainty, but a careful accounting of its presence. Each confidence value is, inevitably, a prediction of future error – a prophecy written in the language of least regret. The system will grow, accruing not just data, but the weight of its own incomplete knowledge.
The true challenge lies not in representing what is known, but in gracefully handling what remains unsaid, unproven, or actively contested. Future iterations will undoubtedly focus on refining these scoring mechanisms, striving for ever-finer granularity. However, it is worth remembering that perfect accuracy is a mirage. Every refinement will reveal new ambiguities, each corrected error birthing a fresh set of unforeseen consequences.
The pursuit of ‘reliable’ question answering is, at its heart, an exercise in controlled forgetting. The system must learn to unlearn, to discard outdated assumptions, and to embrace the inherent messiness of reality. The graph will evolve, branching and pruning, a living record of both discovery and disillusionment. And in that growth, there will be repentance.
Original article: https://arxiv.org/pdf/2601.09720.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- God Of War: Sons Of Sparta – Interactive Map
- Someone Made a SNES-Like Version of Super Mario Bros. Wonder, and You Can Play it for Free
- Epic Games Store Free Games for November 6 Are Great for the Busy Holiday Season
- How to Unlock & Upgrade Hobbies in Heartopia
- One Piece Chapter 1175 Preview, Release Date, And What To Expect
- Battlefield 6 Open Beta Anti-Cheat Has Weird Issue on PC
- Overwatch is Nerfing One of Its New Heroes From Reign of Talon Season 1
- Sony Shuts Down PlayStation Stars Loyalty Program
- The Mandalorian & Grogu Hits A Worrying Star Wars Snag Ahead Of Its Release
- EUR USD PREDICTION
2026-01-18 08:18