Author: Denis Avetisyan
A new algorithm overcomes reconstruction artifacts in digital holographic microscopy, enabling accurate quantitative phase imaging of large tissue samples.
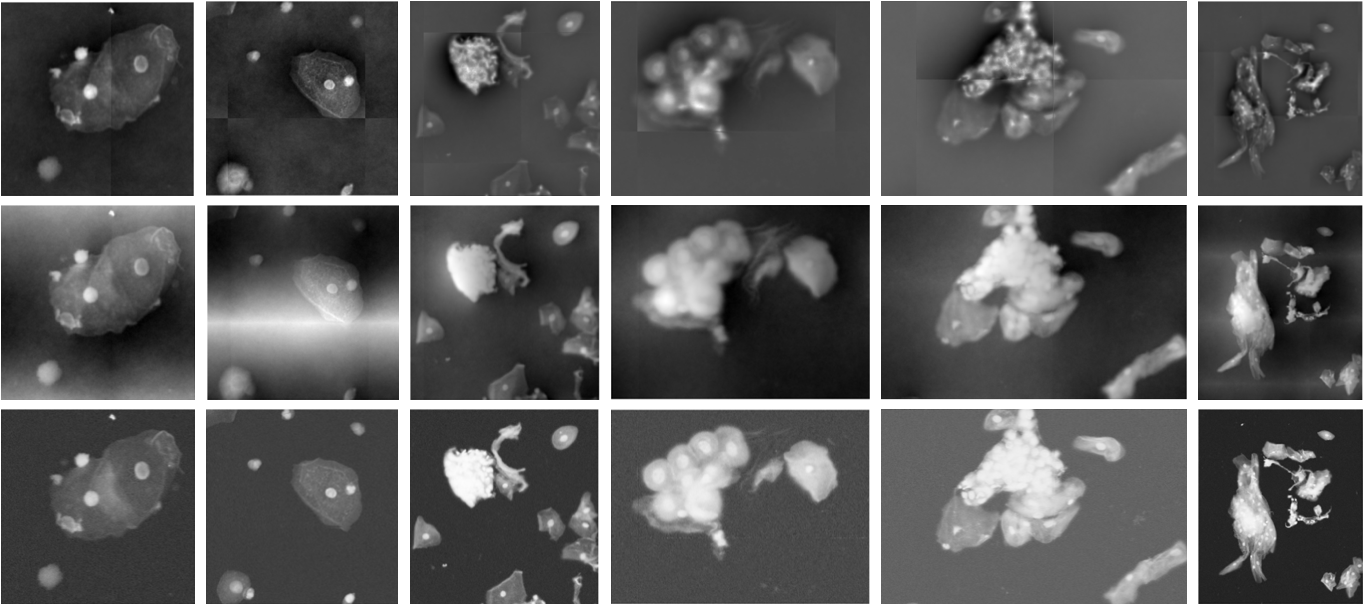
This work presents a method for reconstructing patched digital holograms, improving the accuracy of optical height measurements and enabling high-throughput cell analysis.
Despite advances in both cytological screening via whole slide imaging (WSI) and rich cell characterization through quantitative phase imaging (QPI), these techniques remain largely disjointed. This work, titled ‘Reconstructing Patched or Partial Holograms to allow for Whole Slide Imaging with a Self-Referencing Holographic Microscope’, addresses this gap by introducing a novel reconstruction algorithm capable of generating accurate phase images from digital holograms assembled from multiple patches. The algorithm effectively mitigates artifacts inherent in the WSI patching process, enabling high-throughput analysis of cellular morphology and optical height. Could this approach pave the way for more comprehensive and automated diagnostic tools in fields like cervical cancer screening?
Beyond Qualitative Observation: The Promise of Quantitative Phase Imaging
For over a century, traditional microscopy has served as a cornerstone of biological investigation, providing crucial visual insights into cellular structures. However, this reliance on qualitative observation presents limitations when precise, objective analysis is required. While skilled microscopists can discern morphological changes, quantifying these features-or extracting data regarding cell mass, volume, or dry weight-often relies on subjective assessment or indirect measurements. This lack of quantitative rigor hinders detailed cell analysis, particularly in contexts like disease diagnostics where subtle changes can be indicative of pathology. The inability to directly measure key biophysical properties without the use of potentially disruptive labels means that current techniques frequently fall short of providing the nuanced understanding necessary for accurate and reliable conclusions, creating a demand for more advanced imaging modalities.
Conventional microscopy techniques frequently rely on contrast agents or staining to visualize cellular structures, often disrupting the natural state of the cell and introducing potential artifacts. More fundamentally, these approaches struggle to quantitatively assess key biophysical properties without such interventions. Accurately determining parameters like cell thickness and refractive index – indicators of cell density, dry mass, and internal composition – proves difficult because traditional methods primarily capture intensity variations, not the subtle phase shifts of light that encode this information. This limitation hinders a nuanced understanding of cellular behavior and disease mechanisms, as changes in these biophysical properties often precede detectable morphological alterations and can serve as early biomarkers. Without robust, label-free quantification of these characteristics, researchers face challenges in accurately modeling cellular processes and reliably diagnosing pathological conditions.
Quantitative Phase Imaging (QPI) represents a significant advancement in cellular analysis by providing a means to measure subtle variations in refractive index, effectively visualizing transparent specimens without the need for staining or labeling. This technique allows researchers to quantitatively assess critical cell parameters such as dry mass, volume, and thickness – information often inaccessible with conventional microscopy. However, realizing the full potential of QPI demands sophisticated methodologies for data acquisition and analysis; factors like illumination stability, precise sample positioning, and algorithmic correction for aberrations are crucial. Advanced computational techniques, including adaptive optics and machine learning, are increasingly employed to reconstruct high-resolution phase images and extract meaningful biological insights from the complex data generated, pushing the boundaries of non-invasive cellular characterization and opening new avenues for diagnostics and drug discovery.
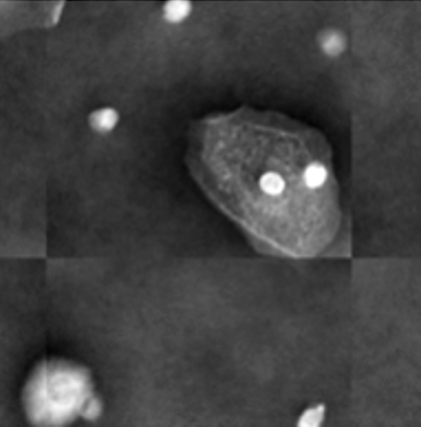
Capturing the Full Wavefront: The Power of Digital Holographic Microscopy
Digital Holographic Microscopy (DHM) distinguishes itself from conventional microscopy by capturing the complete wavefront of light, enabling the recording of both amplitude and phase information of a sample. Traditional microscopy only detects the intensity, or amplitude, of the light, discarding phase data which contains crucial 3D structural information. DHM achieves full wavefront recording through wave interference; a reference beam interferes with the light scattered or transmitted by the sample, creating a hologram. This hologram contains both amplitude and phase encoded as an interference pattern. Subsequent numerical reconstruction of the hologram allows for the retrieval of both amplitude and phase, forming the basis for quantitative phase imaging (QPI) techniques which can provide label-free 3D information about the sample’s refractive index and thickness without the need for physical sectioning or staining.
The Self-Referencing Three-Wave Digital Holographic Microscope (SR-THM) is a specific configuration of Digital Holographic Microscopy designed to maximize the capture of lateral shear. This is achieved through a beam-splitting arrangement where a reference beam is laterally sheared relative to the object beam before interfering at the sensor plane. The resulting hologram contains information about the phase gradient, which directly relates to the object’s refractive index distribution. By precisely controlling and characterizing this lateral shear, the SR-THM enables more accurate and robust phase reconstruction algorithms, minimizing artifacts and improving the quantitative Phase Imaging (QPI) results compared to standard DHM setups. This technique is particularly useful for imaging transparent specimens where phase variations are subtle and require high sensitivity.
Accurate phase reconstruction in Digital Holographic Microscopy (DHM) necessitates the reliable extraction of phase gradients from the recorded interference patterns. This is commonly achieved through the application of the Fourier Transform, a mathematical operation that decomposes the holographic data into its constituent spatial frequencies. Analysis of these frequencies allows for the isolation and quantification of the phase shift introduced by the sample, which is directly proportional to the gradient of the refractive index. Robustness in this process is crucial as errors in phase gradient determination directly translate to inaccuracies in the reconstructed wavefront and subsequent quantitative phase imaging (QPI) results; therefore, advanced algorithms and careful consideration of noise sources are essential for obtaining high-fidelity phase maps.
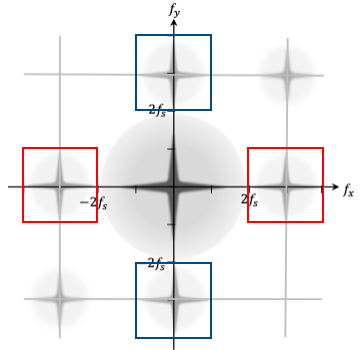
From Gradients to Quantitative Maps: Reconstructing the Phase Landscape
Phase integration is the computational process of determining the quantitative phase distribution from measured phase gradients. Following data acquisition, the phase gradients – representing the rate of phase change – are mathematically integrated to recover the continuous phase values at each point in the sample. This reconstruction is not a simple summation; it requires algorithms to address ambiguities inherent in gradient-based phase recovery. The accuracy of the resulting phase map directly impacts the fidelity of subsequent quantitative analyses, such as the determination of optical height or refractive index variations within the sample. Without effective phase integration, the extracted phase gradients cannot be converted into meaningful, spatially resolved phase information.
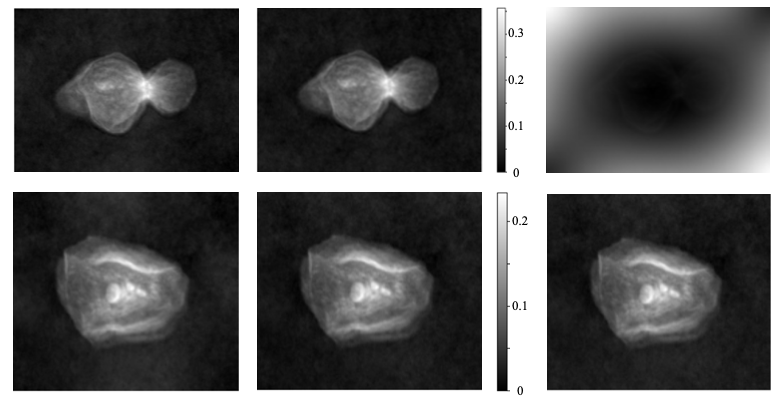
The reconstruction of quantitative phase maps from gradient data is susceptible to artifacts and discontinuities, especially when generating large-area images from tiled or patched data acquisition. Discontinuities arise at patch boundaries due to imperfect alignment or differing gradient estimations, manifesting as artificial steps in the reconstructed phase. These errors propagate during Optical Height calculations and diminish the accuracy of the final quantitative map. Artifacts can also occur from noise amplification during the integration process, requiring careful filtering or weighting strategies. Addressing these issues necessitates algorithms designed to minimize error accumulation and enforce phase continuity across the entire reconstructed image.
Several phase integration methods address artifacts and discontinuities during quantitative phase reconstruction. Iterative-Least Square methods refine phase estimates through iterative adjustments minimizing residual errors. Mirrored Derivative Integration and Antisymmetric Derivative Integration techniques specifically address boundary issues by leveraging derivative information at patch edges, reducing discontinuities. An adapted phase integration method demonstrated improved reconstruction in 157 of 169 cells analyzed, indicating a significant enhancement in phase map accuracy and subsequent Optical Height calculations compared to standard integration approaches.
Validating Reconstruction Fidelity: Ensuring Data Integrity
Accurate quantification of reconstruction error is critical for validating the fidelity of phase map reconstructions. While visual inspection can identify gross errors, objective metrics are necessary for precise evaluation and comparison of reconstruction algorithms. The Normalized L1-Norm calculates the sum of absolute differences between the reconstructed and ground truth phase maps, normalized by the total number of pixels, providing a global error measure. Complementarily, the Mean Difference calculates the average difference between corresponding pixel values, offering insight into systematic biases. These metrics, when applied consistently, enable quantitative assessment of reconstruction quality and facilitate optimization of reconstruction parameters to minimize error and ensure reliable data analysis.
Whole-slide imaging frequently requires the acquisition of images as individual, overlapping patches due to limitations in sensor size and memory capacity. This image patching process, while enabling the digitization of large tissue samples, inherently introduces artifacts at the boundaries between these patches. These artifacts manifest as discontinuities in signal intensity or structural features, potentially leading to inaccurate image analysis and misinterpretation of biological data. Careful consideration must be given to patch overlap, blending algorithms, and subsequent processing steps to minimize the visibility and impact of these boundary artifacts on the final reconstructed image.
Robust integration methods, specifically Mirrored Derivative Integration (MDI), are critical for achieving high-fidelity reconstruction in whole-slide imaging. Analysis demonstrates that MDI significantly reduces local errors during image patching, enabling reconstruction with errors of ≤4 µm. Comparative data indicates a substantial performance difference; without MDI, only 20% of analyzed cells were accurately reconstructed, highlighting the method’s efficacy in improving reconstruction accuracy and overall data quality.
Towards Automated Diagnostics: Unlocking Cellular Insights
Computer-aided diagnostics currently relies heavily on analyzing light intensity, but this approach often overlooks crucial details about a cell’s mass, volume, and refractive index – properties deeply linked to its health and function. Quantitative phase imaging (QPI) addresses this limitation by measuring how light waves change as they pass through a cell, revealing these otherwise invisible characteristics. This provides a far more complete morphological profile than traditional brightfield microscopy, allowing for the detection of subtle changes in cellular structure and density that may precede visible signs of disease. The resulting phase maps, essentially topographical representations of cellular thickness and composition, offer a wealth of information for diagnostic algorithms, promising to significantly enhance the accuracy and speed of disease detection and classification compared to methods based solely on light absorbance.
The advent of deep learning offers a powerful pathway to translate the wealth of information contained within quantitative phase imaging (QPI) data into automated disease diagnostics. These algorithms, specifically convolutional neural networks, are capable of learning complex patterns directly from the phase maps – effectively recognizing subtle morphological alterations in cells that might be imperceptible to the human eye. Training these networks requires large datasets of labeled phase images, but once established, they can rapidly analyze new samples, identifying indicators of disease with remarkable accuracy and consistency. This automated analysis not only accelerates the diagnostic process but also reduces inter-observer variability, providing a more objective assessment of cellular health and potentially enabling earlier disease detection. Furthermore, the algorithms can be refined over time with additional data, continuously improving their performance and expanding their capacity to recognize a wider range of pathological conditions.
The convergence of quantitative phase imaging (QPI) and computer-aided diagnostics (CAD) represents a significant leap forward for cytology, promising to transform disease diagnosis through increased speed, precision, and objectivity. Traditional cytology relies heavily on subjective visual assessment, but QPI offers a means to quantitatively measure cellular properties – specifically, optical height – which are often indicative of pathological changes. Studies have demonstrated measurable optical height ranges within healthy epithelial cells, typically between [0, 0.25 µm], though adapted phase integration techniques can extend this to [0, 0.3 µm], with maximum observed values reaching 0.65 µm. These subtle, yet quantifiable, variations in cellular morphology, when analyzed with deep learning algorithms as part of a CAD system, can reveal early indicators of disease that might otherwise be missed, ultimately leading to more accurate and timely diagnoses.
The presented research directly addresses a critical limitation in whole slide imaging – the introduction of artifacts during the holographic patching process. This pursuit of accurate phase reconstruction, despite technical challenges, echoes John Stuart Mill’s assertion that “It is better to be a dissatisfied Socrates than a satisfied fool.” The algorithm’s focus on minimizing errors and achieving faithful representation of optical height isn’t merely a technical refinement; it’s a commitment to intellectual honesty. Scalability without ethics, in this instance, would manifest as high-throughput analysis built upon flawed data, rendering the entire process unreliable. Only value control – prioritizing data integrity – makes the system truly safe and meaningful for applications like cytological screening.
Beyond the Patch
The reconstruction of whole slide images from fragmented holograms addresses a practical hurdle, but it simultaneously highlights a deeper issue: the relentless push for throughput often outpaces careful consideration of data integrity. This work, while elegantly mitigating patching artifacts, does not erase the fundamental question of what is lost – and what values are silently encoded – in the translation of a complex, three-dimensional sample into a two-dimensional, computationally reconstructed image. Every algorithm has morality, even if silent, and the ease with which data can be ‘fixed’ should not obscure the imperative to minimize distortion at the source.
Future efforts must move beyond simply correcting for technical limitations. The field needs to confront the inherent trade-offs between speed, resolution, and the preservation of biological nuance. Can mirrored derivative integration, or other advanced phase reconstruction techniques, reveal not only what is present in a sample, but also how it is presented – its structural integrity, its subtle variations? Scaling without value checks is a crime against the future.
Ultimately, the true innovation will not lie in clever algorithms, but in a paradigm shift. A future where quantitative phase imaging prioritizes information fidelity over sheer volume, and recognizes that the most powerful insights arise not from processing more data, but from understanding it more deeply. The challenge is to build instruments – and algorithms – that serve biological truth, not merely the demands of automation.
Original article: https://arxiv.org/pdf/2601.15952.pdf
Contact the author: https://www.linkedin.com/in/avetisyan/
See also:
- EUR USD PREDICTION
- TRX PREDICTION. TRX cryptocurrency
- Epic Games Store Free Games for November 6 Are Great for the Busy Holiday Season
- Xbox Game Pass September Wave 1 Revealed
- How to Unlock & Upgrade Hobbies in Heartopia
- Battlefield 6 Open Beta Anti-Cheat Has Weird Issue on PC
- Sony Shuts Down PlayStation Stars Loyalty Program
- The Mandalorian & Grogu Hits A Worrying Star Wars Snag Ahead Of Its Release
- How to Increase Corrosion Resistance in StarRupture
- Best Ship Quest Order in Dragon Quest 2 Remake
2026-01-24 20:36